Lactic acid bacteria obtained from cereal-based fermented food products at different processing stages
Main Article Content
Abstract
Background: Selective consumption of fermented foods obtained from dairy sources have resulted from problems of lactose intolerance, dairy allergies and strict vegetarian dietary habits. Non-dairy, cereal-based fermented food products undergo different processing stages and it is believed that lactic acid bacteria (LAB), is involved in the fermentation process. However, there are little or no information on the specific LAB that are involved at the various intermediate stages.
Objectives: The aim of this study was to explore indigenous cereal food products as an alternative source of LAB as well as isolate and identify LAB from cereal-based fermented food obtained at different processing stages.
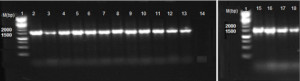
Methods: Two varieties of corn/maize and sorghum (guinea corn) were cleaned, steeped in water and milled. Samples obtained at different processing stages were collected into sterile containers. LAB were isolated on De Man, Rogosa and Sharpe agar and characterized using biochemical, microscopic and molecular methods.
Results: The total viable bacterial cell count ranged from 9.22 to 9.66 log10 CFU/ml. Conventional identification method revealed rod-shaped, Gram positive, catalase negative, non-spore forming bacteria with single, paired and long chain cell arrangements. The 16S rRNA gene sequence analysis identified diverse species of two LAB groups namely: Lactobacillus (93.02%) and Pediococcus (6.98%) with L. fermentum as the dominant Lactobacillus spp.
Conclusion: This study revealed the presence of LAB from fermented maize and sorghum at different processing stages. Our findings show that the slurry-processed-stage, which is the commonly consumed fermented food product apparently contains similar diverse LAB as identified in the milled whole product.
Downloads
Article Details
Issue
Section

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
How to Cite
Share
References
Rezac S, Kok CR, Heermann M, Hutkins R (2018). Fermented Foods as a Dietary Source of Live Organisms. Frontiers in Microbiology 9:1785.
Angelescu IR, Zamfir M, Stancu MM, Grosu-Tudor SS (2019). Identification and probiotic properties of lactobacilli isolated from two different fermented beverages. Annals of Microbiology 69(13):1557-1565.
Achi OK, Asamudo NU (2019). Cereal-BasedFermented Foods of Africa as Functional Foods. In:Mérillon JM, Ramawat KG, eds. Bioactive Molecules in Food. Reference Series in Phytochemistry. Springer International Publishing, pp.1527-1558.
Ranadheera CS, Vidanarachchi JK, Rocha RS, Cruz AG, Ajlouni S (2017). Probiotic Delivery through Fermentation: Dairy vs. Non-Dairy Beverages. Fermentation 3(4):67.
Sornplang P, Piyadeatsoontorn S (2016). Probiotic isolates from unconventional sources: a review. Journal of Animal Science and Technology 58(1):26.
Guyot JP (2012). Cereal-based fermented foods in developing countries: ancient foods for modern research. International Journal of Food & Science Technology 47(6):1109-1114.
Evans E, Musa A, Abubakar Y, Mainuna B (2013). "Nigerian indigenous fermented foods: processes and prospects." Mycotoxin and food safety in developing countries 153:153-180.
Iwuoha CI, Eke OS (1996). Nigerian indigenous fermented foods: their traditional processoperation, inherent problems, improvements and
current status. Food Research International 29(5):527-540.
Afolayan AO, Ayeni FA, Ruppitsch W (2017). Antagonistic and Quantitative Assessment of Indigenous Lactic acid Bacteria in Different Varieties of Ogi against Gastrointestinal Pathogens. The Pan African Medical Journal 27:22.
Shahane AA, Shivay YS (2016). Cereal Residues - Not a Waste Until We Waste it: A Review. International Journal of Bio-resource and Stress Management 7(1):162-173.
Grujovi MŽ, Mladenovi KG, Semedo-Lemsaddek T, Laranjo M, Stefanovi OD, Koci-Tanackov SD (2022). Advantages and disadvantages of non-starter lactic acid bacteria from traditional fermented foods: Potential use as starters or probiotics. Comprehensive Reviews in Food cience and Food Safety 21(2):1537-1567.
Şanlier N, Gökcen BB, Sezgin AC (2019). Health benefits of fermented foods. Critical Review in Food Science and Nutrition 59(3):506-527.
Tamang JP, Watanabe K, Holzapfel WH (2016). Review: Diversity of Microorganisms in Global Fermented Foods and Beverages. Frontiers in
Microbiology 7:377
Papadimitriou K, Alegría Á, Bron PA, de Angelis M, Gobbetti M, Kleerebezem M, Lemos JA, Linares DM, Ross P, Stanton C, Turroni F, van Sinderen D, Varmanen P, Ventura M, Zúñiga M, Tsakalidou E, Kok J (2016). Stress Physiology of Lactic Acid Bacteria. Microbiology and Molecular Biology Reviews 80(3):837-890.
Mokoena MP (2017). Lactic Acid Bacteria and Their Bacteriocins: Classification, Biosynthesis and Applications against Uropathogens: A Mini-Review. Molecules 22(8):1255.
Bintsis T (2018). Lactic acid bacteria as starter cultures: An update in their metabolism and genetics. AIMS Microbiology 4(4):665-684.
Olatunde OO, Obadina AO, Omemu AM, Oyewole OB, Olugbile A, Olukomaiya OO (2018). Screening and molecular identification of potential probiotic lactic acid bacteria in effluents generated during ogi production. Annals of Microbiology 68(7):433-443.
Ijabadeniyi AO (2007). Microorganisms associated with ogi traditionally produced from three varieties of maize. Research Journal of Microbiology 2(3):247- 253.
Tilahun B, Tesfaye A, Muleta D, Bahiru A, Terefework Z, Wessel G (2018). Isolation and Molecular Identification of Lactic Acid Bacteria Using 16s rRNA Genes from Fermented Teff (Eragrostis tef (Zucc.)) Dough. International Journal of Food Science 2018:8510620.
Mulaw G, Sisay Tessema T, Muleta D, Tesfaye A (2017). In Vitro Evaluation of Probiotic Properties of Lactic Acid Bacteria Isolated from Some Traditionally Fermented Ethiopian Food Products. International Journal of Microbiology 2019:7179514.
Cheesbrough M (2005). District Laboratory Practice in Tropical Countries, Part 2. Cambridge University Press.
David AAD, Orukotan AA, Mohammed SSD (2019).Conventional and molecular characterization of selected Lactic acid bacteria from fermented corn gruel (ogi) and fermented milk (nono). Science World Journal 14(4):28-34.
Bergkessel M, Guthrie C (2013). Colony PCR. Methods in Enzymology 529:299-309.
Lane, D.J. (1991) 16S/23S rRNA Sequencing. In: Stackebrandt, E. and Goodfellow, M., Eds., Nucleic 175 West African Journal of Pharmacy (2023) 34 (2) Acid Techniques in Bacterial Systematic, John Wiley and Sons, New York, pp. 115-175.
Turner S, Pryer KM, Miao VPW, Palmer JD (1999). Investigating Deep Phylogenetic Relationships among Cyanobacteria and Plastids by Small Subunit rRNA Sequence Analysis1. Journal of Eukaryotic Microbiology 46(4):327-338.
NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information (2018). Nucleic Acids Research 46(D1):D8-D13.
Dei HK (2017). Assessment of Maize (Zea mays) as Feed Resource for Poultry. Poultry Science 1:1-32.
Capozzi V, Russo P, Dueñas MT, López P, Spano G (2012). Lactic acid bacteria producing B-group vitamins: a great potential for functional cereals products. Applied Microbiology and Biotechnology 96(6):1383-1394.
Liptáková D, Matejčeková Z, Valík Ľ (2017). Lactic Acid Bacteria and Fermentation of Cereals and Pseudocereals. Fermentation Processes 10:65459.
Carrasco G, Valdezate S, Garrido N, Villalón P, Medina-Pascual MJ, Sáez-Nieto JA (2013). Identification, Typing , and Phylogenetic
Relationships of the Main Clinical Nocardia Species in Spain According to Their gyrB and rpoB Genes. Journal of Clinical Microbiology 51(11):3602-3608.
Lee HW, Roh SW, Shin NR, Lee J, Whon TW, Jung MJ, Yun JH, Kim MS, Hyun DW, Kim D, Bae JW (2013). Blastopirellula cremea sp. nov., isolated from a dead ark clam. International Journal of Systematic and Evolutionary Microbiology 63(Pt_6):2314-2319.
Clarridge JE 3rd (2004). Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clinical Microbiology Reviews 17(4):840-862.
Osman MA, Neoh H min, Ab Mutalib NS, Chin SF, Jamal R (2018). 16S rRNA Gene Sequencing for Deciphering the Colorectal Cancer Gut Microbiome: Current Protocols and Workflows. Frontiers in Microbiology 9:767.
McGovern E, Waters SM, Blackshields G, McCabe MS (2018). Evaluating Established Methods for Rumen 16S rRNA Amplicon Sequencing with Mock Microbial Populations. Frontiers in Microbiology 9:1365.
Fouhy F, Clooney AG, Stanton C, Claesson MJ, Cotter PD (2016). 16S rRNA gene sequencing of mock microbial populations- impact of DNA extraction method, primer choice and sequencing platform. BMC Microbiology 16(1):123.
Aseel MK, B. Eltahir H, Hamed Elnil Y, E. Elfaki A (2018). Molecular Characterization of Lactic Acid Bacteria Isolated from Starter Dough of Sudanese Sorghum Fermented Flat Bread (Kissra). Pakistan Journal of Nutrition 17(2):57-63.
Owusu-Kwarteng J, Tano-Debrah K, Akabanda F, Jespersen L (2015). Technological properties and probiotic potential of Lactobacillus fermentum strains isolated from West African fermented millet dough. BMC Microbiology 15(1):261.
Naghmouchi K, Belguesmia Y, Bendali F, Spano G, Seal BS, Drider D (2020). Lactobacillus fermentum: a bacterial species with potential for food preservation and biomedical applications. Critical Reviews in Food Science and Nutrition 60(20):3387- 3399. 176 West African Journal of Pharmacy (2023) 34 (2)
Arasu MV, Al-Dhabi NA, Ilavenil S, Choi KC, Srigopalram S (2016). In vitro importance of probiotic Lactobacillus plantarum related to medical field. Saudi Journal of Biological Sciences 23(1):S6-S10.
Anonymous (2007). Gamma-aminobutyric acid (GABA), Monograph. Alternative Medical Reviews 12(3):274-279.
Cai Y, Kumai S, Ogawa M, Benno Y, Nakase T (1999). Characterization and identification of Pediococcus species isolated from forage crops and their application for silage preparation. Applied Environmental Microbiology 65(7):2901-2906.
Evers T, Millar S (2002). Cereal Grain Structure and Development: Some Implications for Quality. Journal of Cereal Science 36(3):261-284.