Susceptibility profile of bacteria obtained from human oro-dental plaques at the dental clinic of the University of Benin Teaching Hospital, Benin City, Nigeria
Main Article Content
Abstract
Background: The oral cavity harbours a diverse community of anaerobic bacterial; many of which have been implicated in life threatening oro-dental and systemic infections.
Objectives: This study evaluated the antimicrobial susceptibility profile of aerobic and anaerobic bacterial isolates obtained from plaque scrapings in patients who visited the study center for scaling and polishing.
Methods: Plaques were scraped from the tooth surface of each study participants with the aid of a sterile tweezer; into specimen collection bottles containing 5mL of sodium thioglycolate broth. This was transported within 2 hours to the laboratory and then subcultured into appropriate growth media for aerobic and anaerobic incubation. Anaerobiosis was achieved by chemical method (pyrrogalor crystals+ NaOH solution). Gram staining and biochemical tests were used to identify resultant bacterial colonies. Susceptibility profile of bacteria isolates were determined by agar disc diffusion assay.
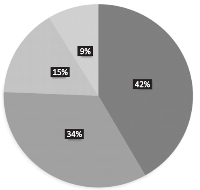
Results: In order of distribution, Streptococcus spp., had the highest frequency of occurrence (42%), followed by Prevotella spp which accounted for (34%). Staphylococcus and Clostridium spps. were least frequently encountered accounting for 15 and 9 % respectively. Susceptibility test result showed that the perfloxacin had high antimicrobial activity, while erythromycin, aminoglycosides and cefotaxim were moderately effective against bacterial isolates. However, amoxicillin and cotrimoxazole showed less activity.
Conclusion: Continuous surveillance studies on the antimicrobial susceptibility profiles of plaque associated bacteria is crucial in identifying emerging resistance patterns. This data can guide healthcare providers in adapting treatment strategies and developing new therapeutic approaches in the management of oro-dental infections.
Downloads
Article Details
Issue
Section

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
How to Cite
Share
References
Jakubovics NS, Goodman SD, Mashburn?Warren L, Stafford GP, Cieplik F (2021). The dental plaque biofilm matrix. Periodontology 2000. ;86(1):32-56.
Del Giudice C, Vaia E, Liccardo D, Marzano F, Valletta A, Spagnuolo G, Ferrara N, Rengo C, Cannavo A, Rengo G.(2021). Infective endocarditis: a focus on oral microbiota. Microorganisms.4; 9(6): 1218.
Sedghi L, DiMassa V, Harrington A, Lynch SV, Kapila YL. (2021). the oral microbiome: Role of key organisms and complex networks in oral health and disease. Periodontology 2000. 87(1):107-31.
Warrier A, Satyamoorthy K, Murali TS(2021).Quorum-sensing regulation of virulence factors in bacterial biofilm. Future microbiology. 16(13):1003-21.
McManus MC (1997). Mechanisms of bacterial resistance to antimicrobial agents. American Journal of Health-System Pharmacy.
;54(12):1420-33.
Paniker JA (2006). Ananthanarayan and Paniker's Test book of Microbiology. Orient Blackswan Hyderabad 500 029 (AP) Indian. Pp.39-51.
Paniker JA (2006). Ananthanarayan and Paniker's Test book of Microbiology. Orient Blackswan Hyderabad 500 029 (AP) Indian. Pp.39-51.
Bauer AW, Kirby WN, Sherris JC and Turch M (1966). Antibiotic susceptibility testing by a standardized single disc method. American Journal of Clinical Pathology 45: 493-496.
EUCAST (2015). Breakpoint tables for interpretation of MICs and zone diameters European committee for antimicrobial susceptibility testing (EUCAST) Sweden, 2007, 2005.Version 7.1.
Nobbs AH, Lamont RJ, Jenkinson HF (2009).Streptococcus adherence and colonization. Microbiology and molecular biology reviews. 73(3):407-50.
Jakubovics NS, Yassin SA, Rickard AH (2014). Community interactions of oral streptococci.Advances in Applied Microbiology. 1;87:43-110.
Tognetti L, Martinelli C, Berti S, Hercogova J, Lotti T, Leoncini F, Moretti S (2012).Bacterial skin and soft tissue infections: review of the epidemiology, microbiology, aetiopathogenesis and treatment: a collaboration between dermatologists and infectivologists. Journal of the European Academy of Dermatology and Venereology. 26(8):931-41.
Claeys KC, Hopkins TL, Vega AD, Heil EL (2018). Fluoroquinolone restriction as an effective antimicrobial stewardship intervention. Current
Infectious Disease Reports.20:1-7
Yao JD, Moellering Jr RC. (2011).Antibacterial agents. Manual of Clinical Microbiology. 16:1041-81.
Poole K (2015). Efflux-mediated antimicrobial resistance. Journal of Antimicrobial Chemotherapy. 1;56(1):20-51.
Jelić D, Antolović R(2016). From erythromycin to azithromycin and new potential ribosome-binding antimicrobials. Antibiotics. 1;5(3):29.
Soares GM, Figueiredo LC, Faveri M, Cortelli SC, Duarte PM, Feres M(2012).Mechanisms of bacterial resistance to these drugs. Journal of Applied Oral Science. 20:295-307.
Amsden GW (2001). Advanced-generation mac rolides: tissue-direc ted antibioti c s. International Journal of Antimicrobial Agents. 1;
:11-5.
Takahashi Y, Igarashi M (2018). Destination of aminoglycoside antibiotics in the 'post-antibiotic era'. The Journal of Antibiotics.71 (1):4-14.